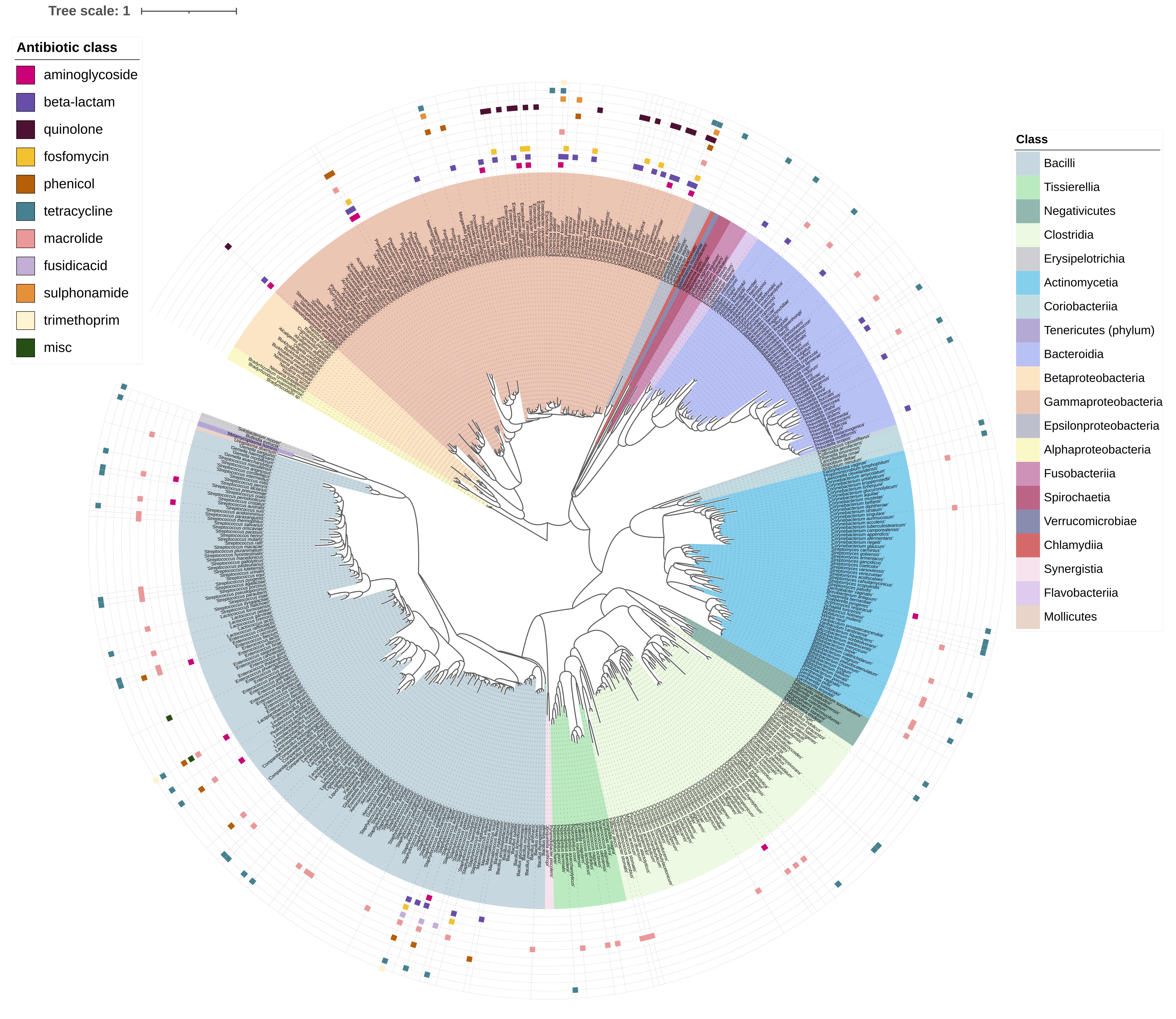
Supplementary Figure 4.
A 16S rDNA-based tree of vaginal microbial species annotated with associated antibiotic resistance classes. Tree coloring indicates bacterial order (legend, upper right). For this representation, all species-ARG pairs within a single sample were displayed. ARGs (legend, left) are displayed along concentric arcs by antibiotic class.
View interactive tree on iTOL
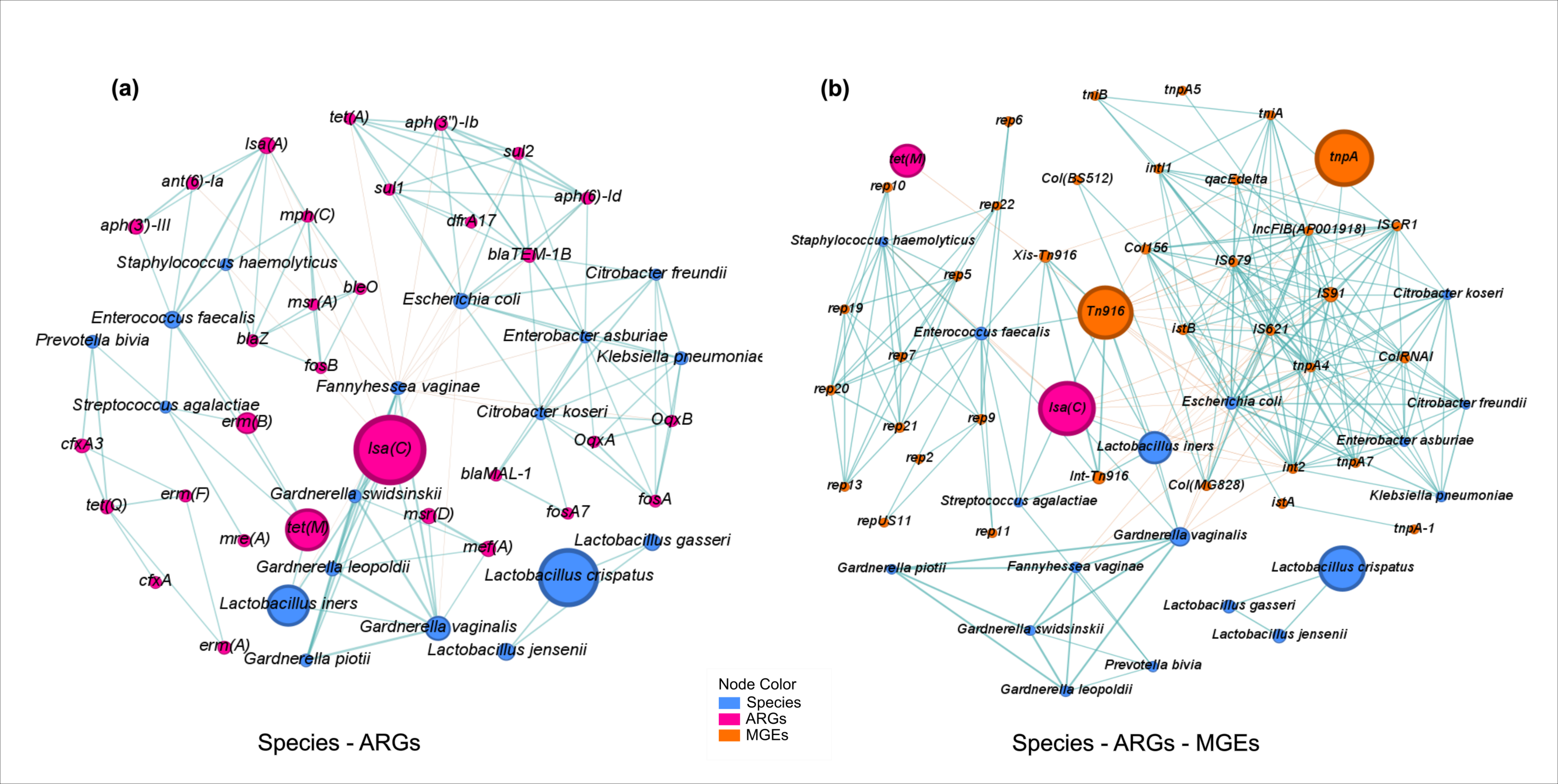
Supplementary Figure 9.
(a) Network constructed from the 20 most abundant microbial species and 50 clinically relevant antimicrobial resistance genes (ARGs). (b) Network including the same 20 microbial species and ARGs with the addition of MGEs. Nodes represent microbial species (blue), ARGs (pink), and MGEs (orange), with node size scaled by average relative abundance (%). Edges indicate significant correlations: blue edges represent positive correlations (ρ > 0.2, P < 0.01), and orange edges represent negative correlations (ρ < -0.2, P < 0.01); edge thickness is proportional to correlation strength.
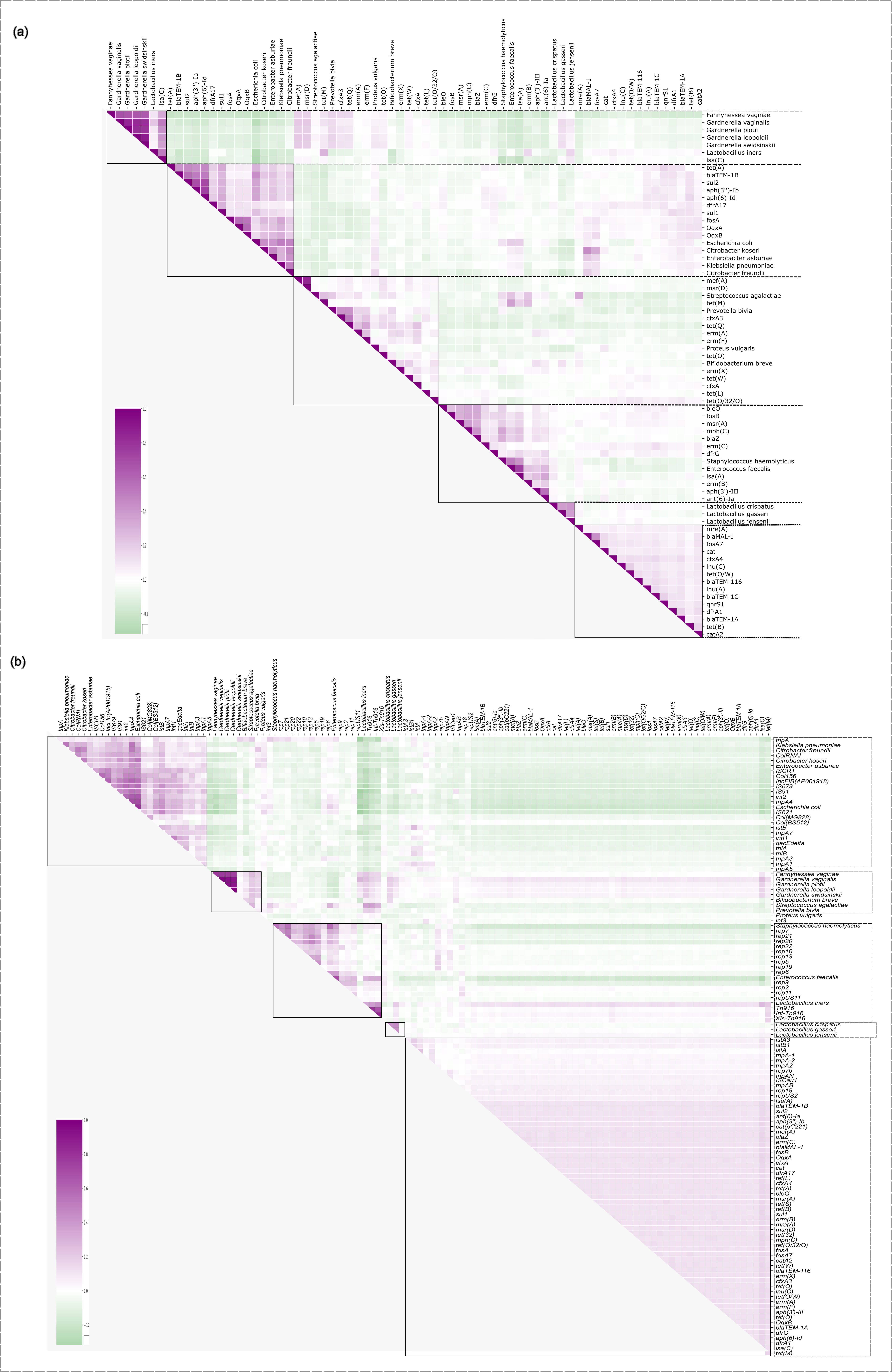
Supplementary Figure 10.
(a) SparCC-based correlation matrix between the 20 most abundant microbial species and the 50 most abundant antibiotic resistance genes (ARGs). (b) SparCC-based correlation matrix beetween the same microbial species and the 50 moste abundant ARGs and mobile genetic elements (MGEs). Matrix was clustered with the “average” linkage method.